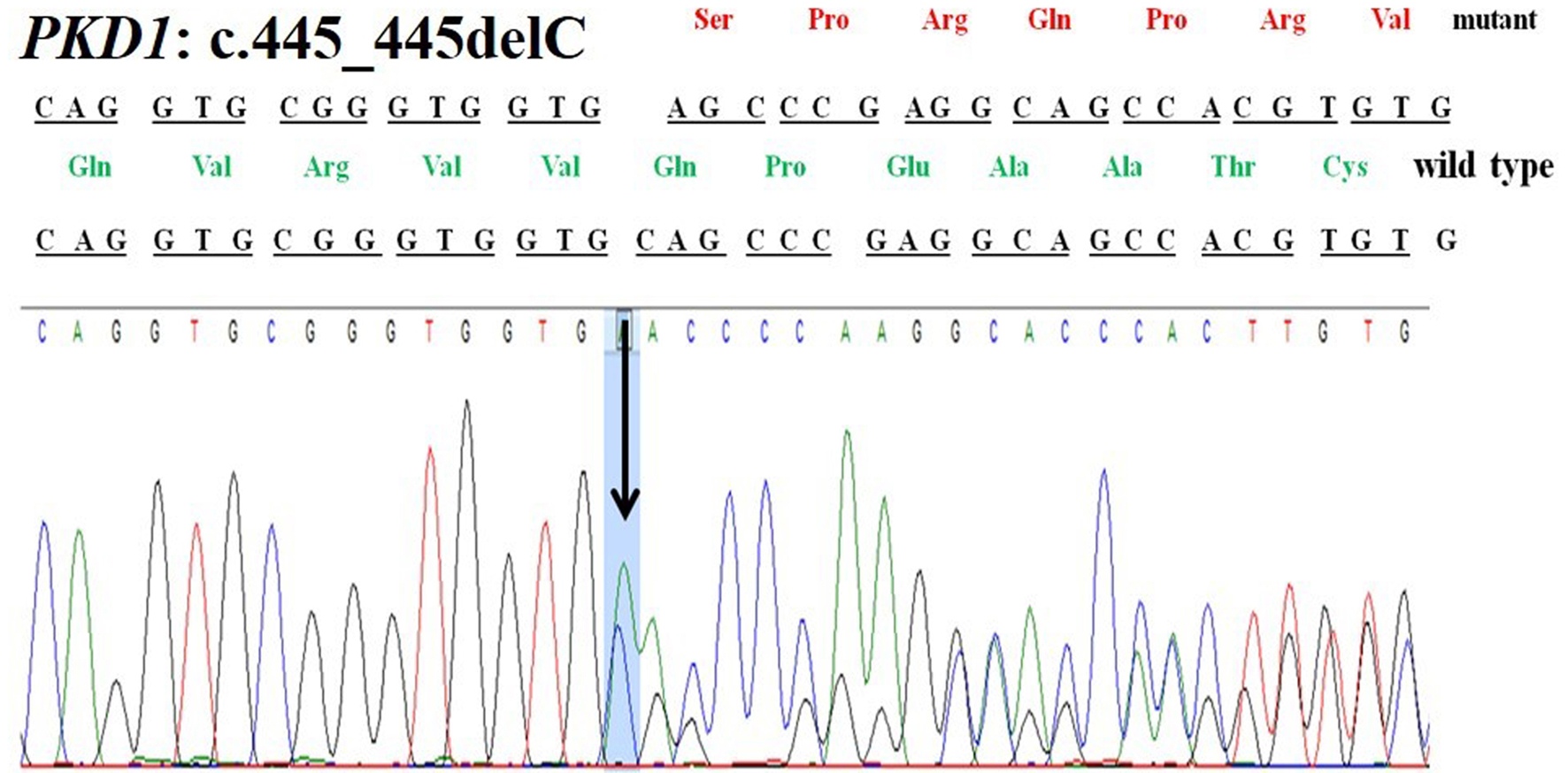
Figure 1. Chromatogram of PKD1: c.445_445delC deletion in patient 16.1.
| World Journal of Nephrology and Urology, ISSN 1927-1239 print, 1927-1247 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Nephrol Urol and Elmer Press Inc |
| Journal website http://www.wjnu.org |
Original Article
Volume 9, Number 1, March 2020, pages 15-24
Characterization of Novel Truncated Variants Identified From Indian Autosomal Dominant Polycystic Kidney Disease Cases
Figures


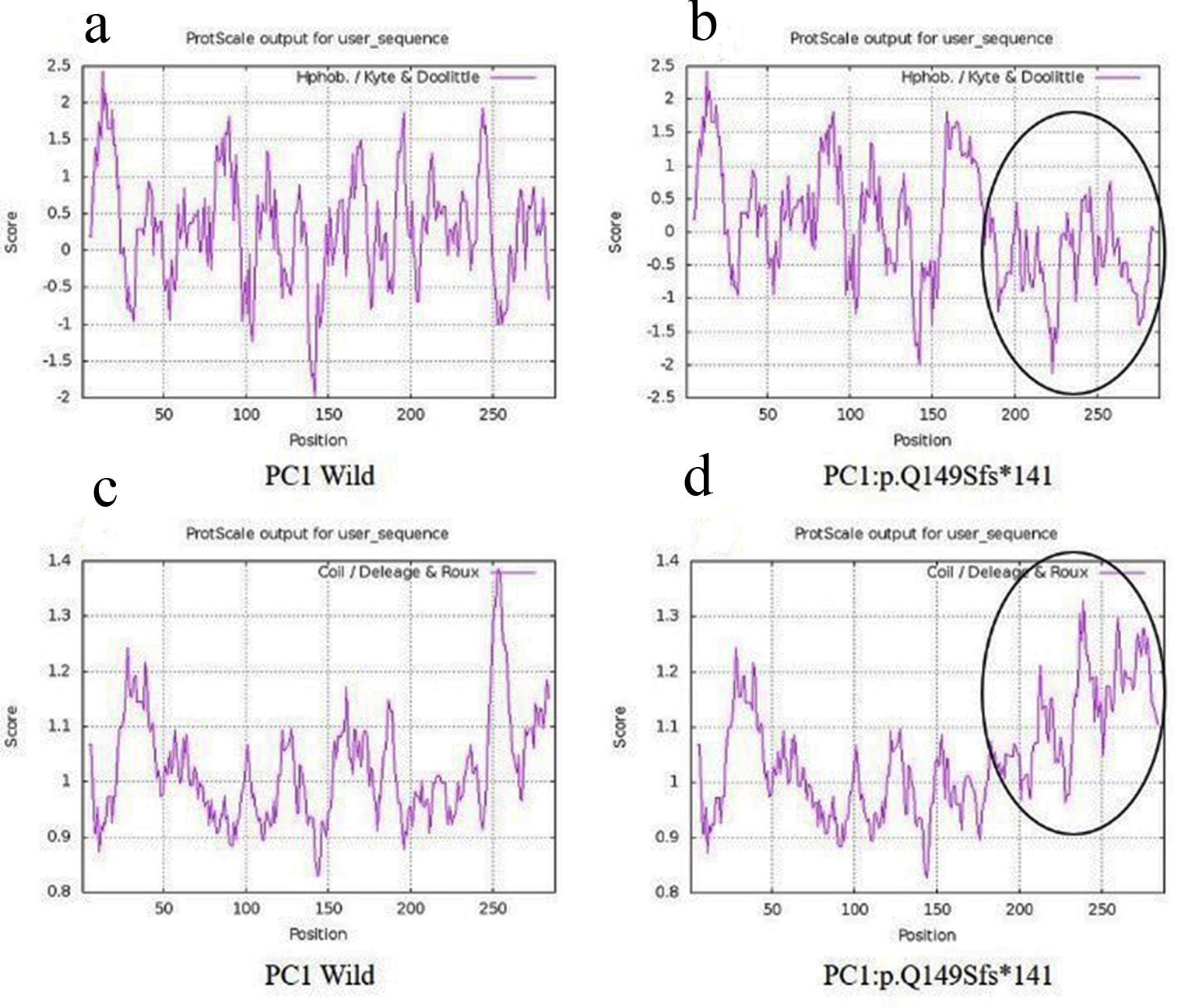
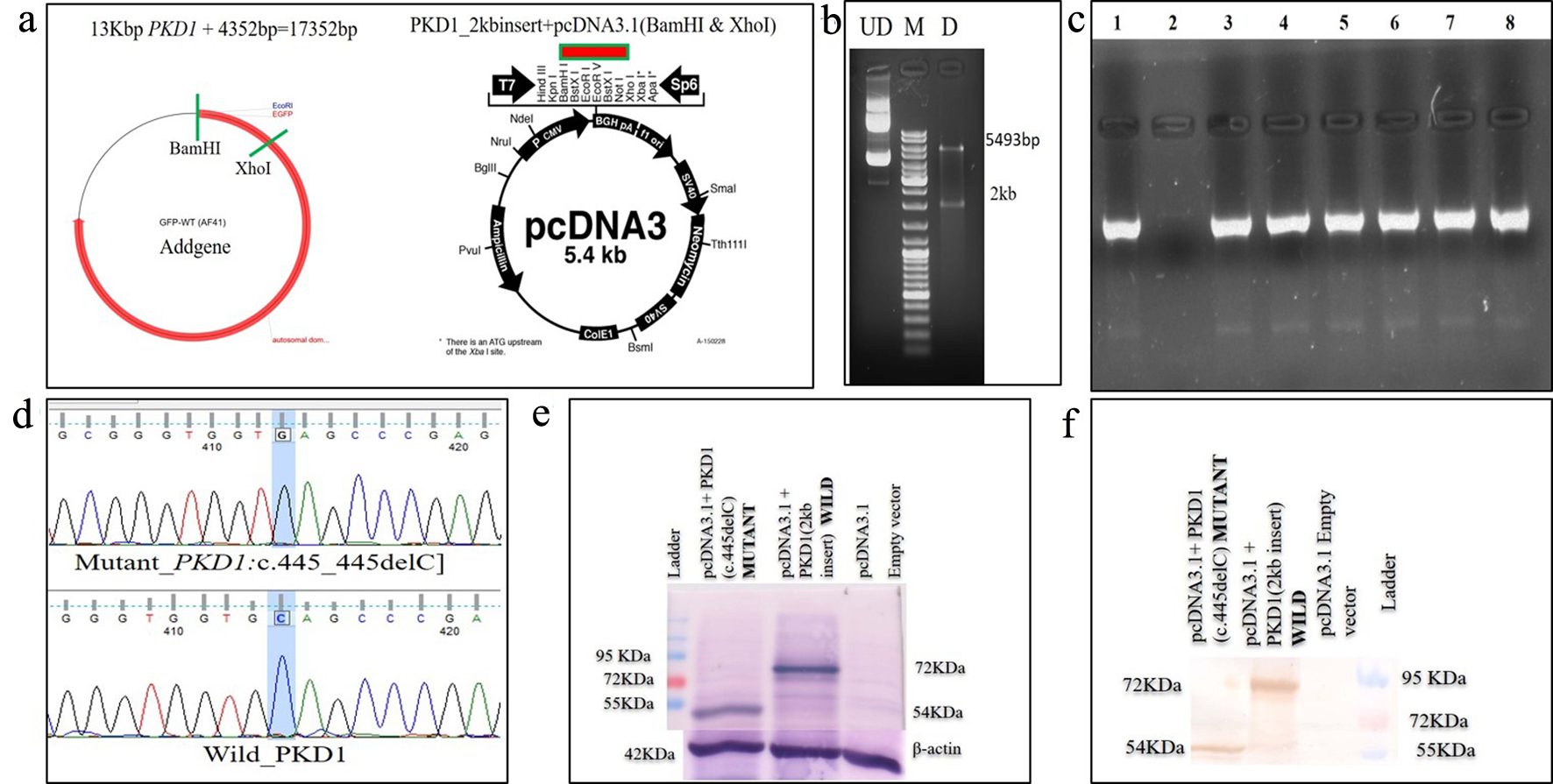
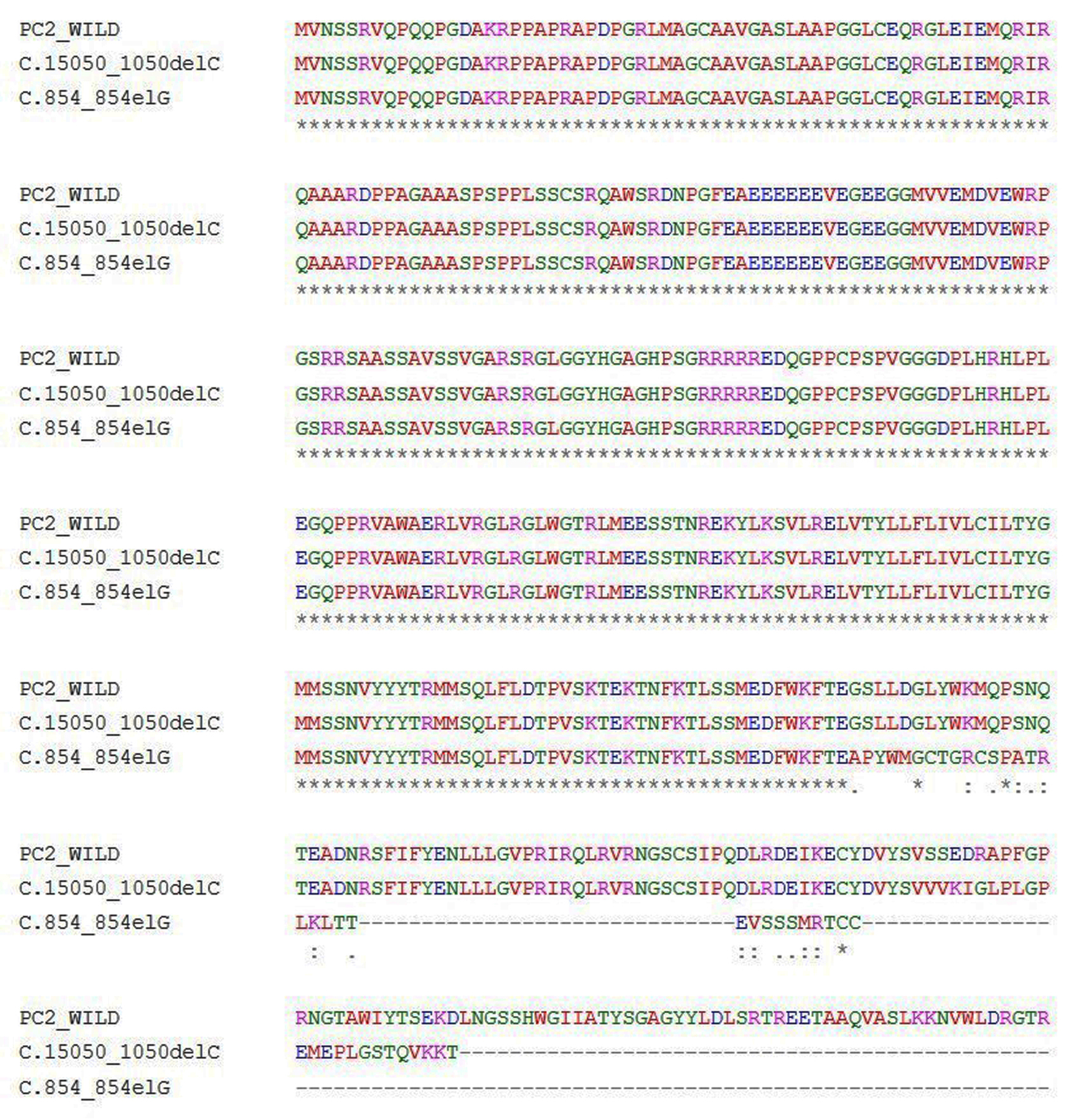
Tables
| Sample ID | PKD1/PKD2 | rs ID | Protein | Domain | In silico | Remark | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation taster | Align-GVGD | PolyPhen2 | PROVEAN | SIFT | PKDB | ||||||
| ND: not defined; NA: not possible to analyze; Align-GVGD: Align-Grantham Variation Grantham Deviation; PolyPhen2: Polymorphism Phenotyping v2; PROVEAN: Protein Variation Effect Analyzer; SIFT: Sorting Intolerant From Tolerant; PKDB: Polycystic Kidney Disease Mutation Database. | |||||||||||
| PKD16.1 | c.445_445delC | NA | p.Q149Sfs*141 | LRRCT | Pathogenic | NA | NA | NA | NA | NA | Disease causing |
| c.7165T>C | rs2457533 | p.L2389L | REJ | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| c.7441C>T | rs2003782 | p.L2481L | REJ | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| c.12133A>G | rs10906 | p.I4045V | TM10 | Polymorphism | Class C25 | Benign | Neutral | Tolerated | Likely neutral | Likely neutral | |
| c.12176C>T | rs3209986 | p.A4059V | ND | Polymorphism | Class C55 | Benign | Neutral | Tolerated | Likely neutral | Likely neutral | |
| c.12276A>G | rs3087632 | p.A4092A | TM11 | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| c.12409C>T | rs79899502 | p.L4137L | ND | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| c.12630T>C | rs7203729 | p.P4210P | ND | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| IVS3-22G>A | rs2725221 | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |||
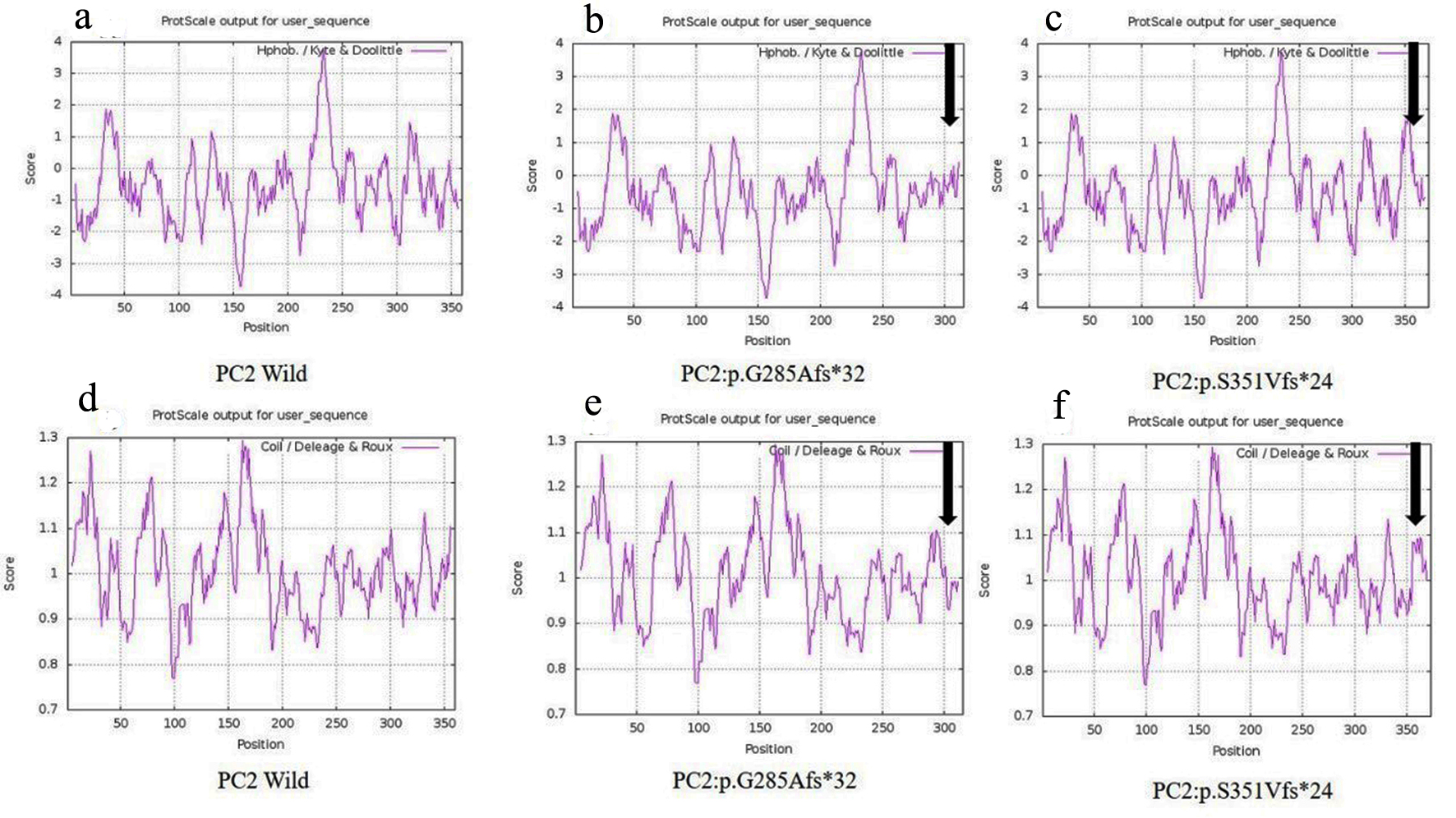
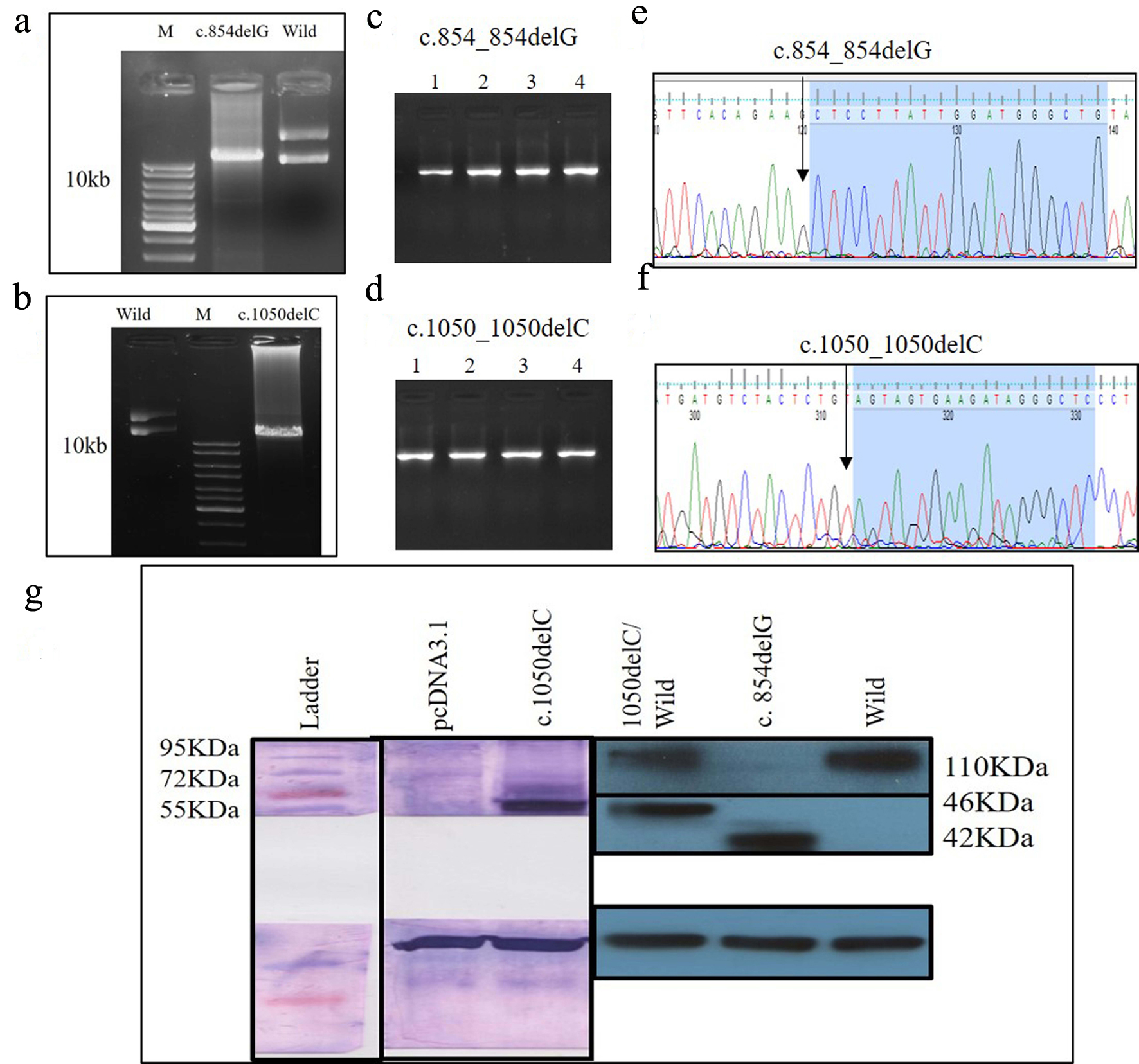
| PKD39.1 | PKD2: c.854_854delG | Reference | p.G285Afs*32 | First extracellular loop | Pathogenic | NA | NA | NA | NA | NA | Disease causing |
| c.1119C>T | rs199685642 | p.L373L | ND | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| PKD64.1 | PKD2: c.1050_1050delC | Reference | p.S351Vfs*24 | First extracellular loop | Pathogenic | NA | NA | NA | NA | NA | Disease causing |
| c.1119C>T | rs199685642 | p.L373L | ND | Polymorphism | NA | NA | NA | NA | Likely neutral | Likely neutral | |
| c.10529C>T | rs45478794 | p.T3510M | ND | Polymorphism | Class C65 | Possibly damaging | Neutral | Damaging | Likely neutral | Damaging | |
| Primer name | Sequence 5′-3′ |
|---|---|
| aPrimers used in sub-cloning; bSDM primers; cSite of nucleotide deletion. | |
| PKD1_F_BamHla | TAATATGGATCCGGCATGGTGAGCAAGGG |
| PKD1_R_Xhola | TAATATCTCGAGGTAGCAGTGCCCGTTGCC |
| PKD1_EX4_C.445delC_PFb | cAGCCCGAGGCAGCCACGT |
| PKD1_EX4_C.445delC_PRb | CACCACCCGCACCTGCTG |
| PKD2_EX4_C.854delC_PFb | cCTCCTTATTGGATGGGCTG |
| PKD2_EX4_C.854delC_PRb | CTTCTGTGAACTTCCAGAAG |
| PKD2_EX4_C.1050delC_PFb | cAGTAGTGAAGATAGGGCTC |
| PKD2_EX4_C.1050delC_PRb | ACAGAGTAGACATCATAGC |